A preliminary study of the effect of room temperature incubation on phylogenetic composition of salivary microbiota
September 26, 2018 / Categories: Digital Dentistry, Implant Dentistry

Enomoto, Atsuko

Hara, Tomoya

Kusano, Kaoru

Yamada, Yoichi

Botticelli, Daniele

Baba, Shunsuke

Nambu, Takayuki

Mashimo, Chiho

Taniguchi, Makoto

Nakatani, Kensuke

Komuro, Akira

Nakajima, Yasushi

Honda, Masaaki

Ito, Yusaku
Abstract
Objective
Human oral microbiome research has revealed that differences in microbial communities may lead to not only oral disease but also systemic conditions. Although large-scale cohort studies are needed to further investigate these relationships, little is known about the storage method of salivary samples collected in the clinical field. In this study, the temporal stability of salivary microbiota at room temperature was investigated to support future cohort studies of the oral microbiome.
Materials and methods
Three salivary samples collected from 2 healthy adults were incubated at 20°C for 3 or 6 h, and the bacterial composition was then analyzed using next-generation sequencing of the 16S ribosomal RNA gene.
Results
Although slight changes in the microbial profiles were observed in the samples incubated for 6 h, principal coordinate analysis revealed significant clustering based on the origin of the salivary samples. In contrast, several increased or decreased operational taxonomic units were also detected under this storage condition.
Conclusion
Within the limitation of the sample size in this preliminary study, it can be stated that room temperature storage of salivary samples for a few hours may not affect the overall microbiota structure, while there are some changes in specific bacterial taxa.
Keywords
Salivary microbiota; room temperature storage; next-generation sequencing.
Introduction
Many bacteria indigenously inhabit the human body surface and digestive tract and maintain symbiotic relationships with humans.1Sender R, Fuchs S, Milo R. Are we really vastly outnumbered? Revisiting the ratio of bacterial to host cells in humans. → Cell. 2016 Jan;164(3):337–40.,2Thomas S, Izard J, Walsh E, Batich K, Chongsathidkiet P, Clarke G, Sela DA, Muller AJ, Mullin JM, Albert K, Gilligan JP, DiGuilio K, Dilbarova R, Alexander W, Prendergast GC. The host microbiome regulates and maintains human health: a primer and perspective for non-microbiologists. → Cancer Res. 2017 Apr;77(8):1783–812. For example, in the intestine, in which the largest number of indigenous bacteria is present, there are about 38 trillion bacteria, comprised of at least 160 species that maintain a symbiotic balance. It has been suggested that an abnormal composition ratio (dysbiosis) of these intestinal bacteria is associated with obesity, type 2 diabetes, autism and allergic disease, in addition to the induction of inflammatory disease.3Schroeder BO, Bäckhed F. Signals from the gut microbiota to distant organs in physiology and disease. → Nat Med. 2016 Oct;22(10):1079–89.,4Hong SW, Kim KS, Surh CD. Beyond hygiene: commensal microbiota and allergic diseases. → Immune Netw. 2017 Feb;17(1):48–59.
The oral cavity is also a niche for indigenous bacterial inhabitation, in which 5–10 billion indigenous bacteria, comprised of about 200 species, are present in adults, and the species and ratio of bacteria constituting it markedly vary among individuals, similar to intestinal indigenous bacteria.5Krishnan K, Chen T, Paster BJ. A practical guide to the oral microbiome and its relation to health and disease. → Oral Dis. 2017 Apr;23(3):276–86. In addition, some oral flora–constituting bacteria have been determined to be not only involved in oral diseases, such as dental caries, periodontal disease and periimplantitis, but also closely affecting the systemic health state, such as the blood pressure, in addition to health of the oral cavity.6Krishnan K, Chen T, Paster BJ. A practical guide to the oral microbiome and its relation to health and disease. → Oral Dis. 2017 Apr;23(3):276–86.,7Koch CD, Gladwin MT, Freeman BA, Lundberg JO, Weitzberg E, Morris A. Enterosalivary nitrate metabolism and the microbiome: intersection of microbial metabolism, nitric oxide and diet in cardiac and pulmonary vascular health. → Free Radic Biol Med. 2017 Apr;105:48–67.
Marked progression of the nucleotide sequencing technique has markedly contributed to the recent, rapid accumulation of knowledge concerning the relationship between bacterial floras and the state of health described. The previous mainstream of sequencing, the Sanger method, takes time and is labor- and cost-intensive for analysis, and the resolution of the obtained data is not very high.8Aas JA, Paster BJ, Stokes LN, Olsen I, Dewhirst FE. Defining the normal bacterial flora of the oral cavity. → J Clin Microbiol. 2005 Nov;43(11):5721–32. Several next-generation sequencers have recently been developed and enabled the acquisition of an enormous amount of base sequence information within a relatively short time, facilitating relatively simple identification of the constitution of oral bacteria at a high analytical resolution, which was previously difficult.9Zaura E. Next-generation sequencing approaches to understanding the oral microbiome. → Adv Dent Res. 2012 Sep;24(2):81–5. Large-scale analysis of oral floras and the development of test and treatment methods targeting oral bacteria are expected, but the procedures have not yet been standardized in some steps because these are novel analytical methods. Firstly, the method to extract DNA from oral samples has not been standardized. This is one of the most important steps of flora analysis using a next-generation sequencer, and biases in the flora data depending on the extraction method used have been reported.10Yamagishi J, Sato Y, Shinozaki N, Ye B, Tsuboi A, Nagasaki M, Yamashita R. Comparison of boiling and robotics automation method in DNA extraction for metagenomic sequencing of human oral microbes. → PLoS One. 2016 Apr;11(4):e0154389.11Abusleme L, Hong BY, Dupuy AK, Strausbaugh LD, Diaz PI. Influence of DNA extraction on oral microbial profiles obtained via 16S rRNA gene sequencing. → J Oral Microbiol. 2014 Apr 23;6. doi: 10.3402/jom. v6.23990.,12Lazarevic V, Gaïa N, Girard M, François P, Schrenzel J. Comparison of DNA extraction methods in analysis of salivary bacterial communities. → PLoS One. 2013 Jul 3;8(7):e67699. doi: 10.1371/journal.pone.0067699.,13Vesty A, Biswas K, Taylor MW, Gear K, Douglas RG. Evaluating the impact of DNA extraction method on the representation of human oral bacterial and fungal communities. → PLoS One. 2017 Jan 18;12(1):e0169877. doi: 10.1371/journal.pone.0169877. To solve this problem, it is essential to employ the same DNA extraction method when comparing samples of flora. Another step that has not been standardized is the method to store collected samples, for which less is known. When it is not possible to extract nucleic acid immediately after sample collection, freezing of the sample at −80°C or lower is the optimal storage method,14Abusleme L, Hong BY, Dupuy AK, Strausbaugh LD, Diaz PI. Influence of DNA extraction on oral microbial profiles obtained via 16S rRNA gene sequencing. → J Oral Microbiol. 2014 Apr 23;6. doi: 10.3402/jom. v6.23990.,15Luo T, Srinivasan U, Ramadugu K, Shedden KA, Neiswanger K, Trumble E, Li JJ, McNeil DW, Crout RJ, Weyant RJ, Marazita ML, Foxman B. Effects of specimen collection methodologies and storage conditions on the short-term stability of oral microbiome taxonomy. → Appl Environ Microbiol. 2016 Sep;82(18):5519–29.,16Kim D, Hofstaedter CE, Zhao C, Mattei L, Tanes C, Clarke E, Lauder A, Sherrill-Mix S, Chehoud C, Kelsen J, Conrad M, Collman RG, Baldassano R, Bushman FD, Bittinger K. Optimizing methods and dodging pitfalls in microbiome research. → Microbiome. 2017 May 5;5(1):52. doi: 10.1186/s40168-017-0267-5.,17Tedjo DI, Jonkers DM, Savelkoul PH, Masclee AA, van Best N, Pierik MJ, Penders J. The effect of sampling and storage on the fecal microbiota composition in healthy and diseased subjects. → PLoS One. 2015 May 29;10(5):e0126685. doi: 10.1371/journal.pone.0126685. but sample storage by rapid freezing is not always possible at clinical sites and in cohort studies. Several effective methods other than deep-freezing, such as the addition of ethanol to samples, have been investigated in the research field of intestinal flora,18Kim D, Hofstaedter CE, Zhao C, Mattei L, Tanes C, Clarke E, Lauder A, Sherrill-Mix S, Chehoud C, Kelsen J, Conrad M, Collman RG, Baldassano R, Bushman FD, Bittinger K. Optimizing methods and dodging pitfalls in microbiome research. → Microbiome. 2017 May 5;5(1):52. doi: 10.1186/s40168-017-0267-5. but to our knowledge, no study has been reported for oral samples other than an experiment on storage for several days using a special medium.19Luo T, Srinivasan U, Ramadugu K, Shedden KA, Neiswanger K, Trumble E, Li JJ, McNeil DW, Crout RJ, Weyant RJ, Marazita ML, Foxman B. Effects of specimen collection methodologies and storage conditions on the short-term stability of oral microbiome taxonomy. → Appl Environ Microbiol. 2016 Sep;82(18):5519–29.
The aim of the present study was to analyze and evaluate changes in floras in oral samples during storage at room temperature for several hours, aiming at securing the reliability of data collected in clinical and cohort studies on the oral flora, which are expected to be frequently performed in the future.
Materials and methods
Salivary sample collection and storage conditions
In order to investigate the storage reliability of salivary samples at room temperature, fresh saliva was incubated at 20°C, and the temporal stability of microbiota composition was monitored for 3 or 6 h. Two samples were collected from the same subject 1 week apart. In our preliminary study, in total 3 salivary samples prepared from 2 healthy individuals were used. DNA from 9 salivary aliquots was successfully extracted, amplified and sequenced.
The study protocol was approved by the Osaka Dental University Medical Ethics Committee (approval No. 110854), and the 2 subjects signed informed consent forms.
The inclusion criterion was age > 20 years. Exclusion criteria were as follows: self-reported presence of periodontitis or dental caries, current daily smoking, and treatment with local or systemic antibiotics within the past 3 months prior to participation. The primary outcome was within-sample change in salivary microbiota composition (assessed by principal coordinate analysis) during storage at room temperature for several hours.
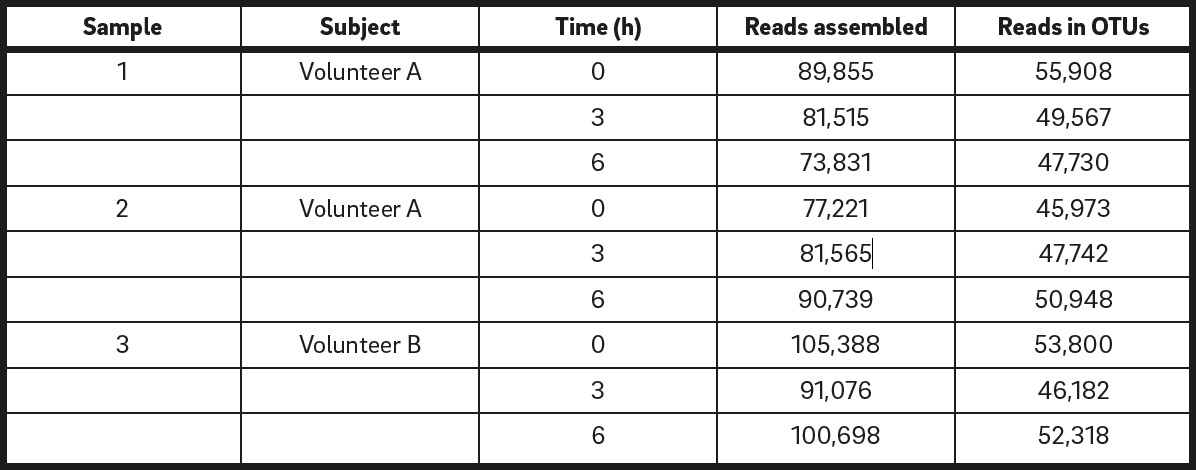
Number of reads assembled and used in OTU generation.
Subjects refrained from eating and drinking for at least 1 h before saliva collection. Three milliliters of saliva was collected in sterile plastic tubes and then homogenized by repetitive pipetting through a narrow bone tip. Aliquots (0.5 mL) of samples were dispensed into 3 sterile tubes, and 1 aliquot was then immediately centrifuged at 10,000 × g for 5 min. The remaining 2 aliquots were incubated at 20°C for 3 and 6 h, respectively, before centrifugation. Next, the supernatant was discarded, and the pellet was stored at -20°C. DNA extraction was carried out within 1 week.
DNA extraction from salivary samples
Bacterial DNA was extracted from 9 salivary samples via chemical and mechanical lysis using a QIAamp UCP Pathogen Mini Kit (Qiagen). The thawed pellets were immediately suspended in 500 μL of ATL buffer containing optional DX reagent, transferred to a Pathogen Lysis Tube S (Qiagen), and then homogenized with Mixer Mill MM 301 (Retsch) for 3 min at a vibrational frequency of 30 Hz. The manufacturer’s protocol was followed thereafter to complete the DNA purification, and DNA was eluted with 50 μL of AVE buffer. After recovery, DNA was also quantified using a Quantus fluorometer (Promega) and a Qubit dsDNA BR Assay Kit (Thermo Fisher Scientific), and stored at –80°C until use. Data collectors were blinded to subject identification.
Library construction and Illumina MiSeq sequencing
Bacterial 16S ribosomal DNA amplification and library construction were performed according to the 16S Metagenomic Sequencing Library Preparation guide supplied by Illumina (part No. 15044223_B). Briefly, the V3–V4 region of the 16S ribosomal RNA gene was amplified via polymerase chain reaction (PCR) using 341f/806r primers and Premix Ex Taq polymerase (Takara Bio). The PCR amplicons were purified using AMPure XP beads (Beckman Coulter). Sequencing adapters containing 8 bp indices were incorporated into the 3′ and 5′ ends of the purified amplicons by PCR. The PCR amplicons were again purified with AMPure XP beads and quantified using a Quantus fluorometer and a Qubit dsDNA HS Assay Kit (Life Technologies). After pooling equimolar amounts of amplicons, 5% equimolar of PhiX DNA (Illumina) was added, and the final library was paired-end sequenced at 2 × 250 bp using a MiSeq Reagent Kit v2 on the Illumina MiSeq platform. Sequencing was performed at the Oral Microbiome Center in Takamatsu, Japan.
Data analysis
The sequences were analyzed with CLC Workbench software (Version 10.0.1; Filgen) equipped with the Microbial Genomics Module plugin (Version 2.0; Filgen). Quality and chimera crossover filtering were performed using default parameters. Operational taxonomic unit (OTU) clustering and taxonomic assignment were carried out using the Human Oral Microbiome Database (Version 14.51) as reference (clustered at 97%). Low-abundance OTUs were discarded from further analyses (less than10 reads, less than 0.01% relative abundance). Alpha diversity was calculated using the number of OTUs. Principal coordinate analysis (PCoA) was performed on the weighted UniFrac matrix, as indicated by the significance of clustering determined by PERMANOVA with 9,999 permutations. PCoA plots were generated by the R package (Version 3.3.3; R Foundation for Statistical Computing, Vienna, Austria. URL http://www.R-project.org/).
-
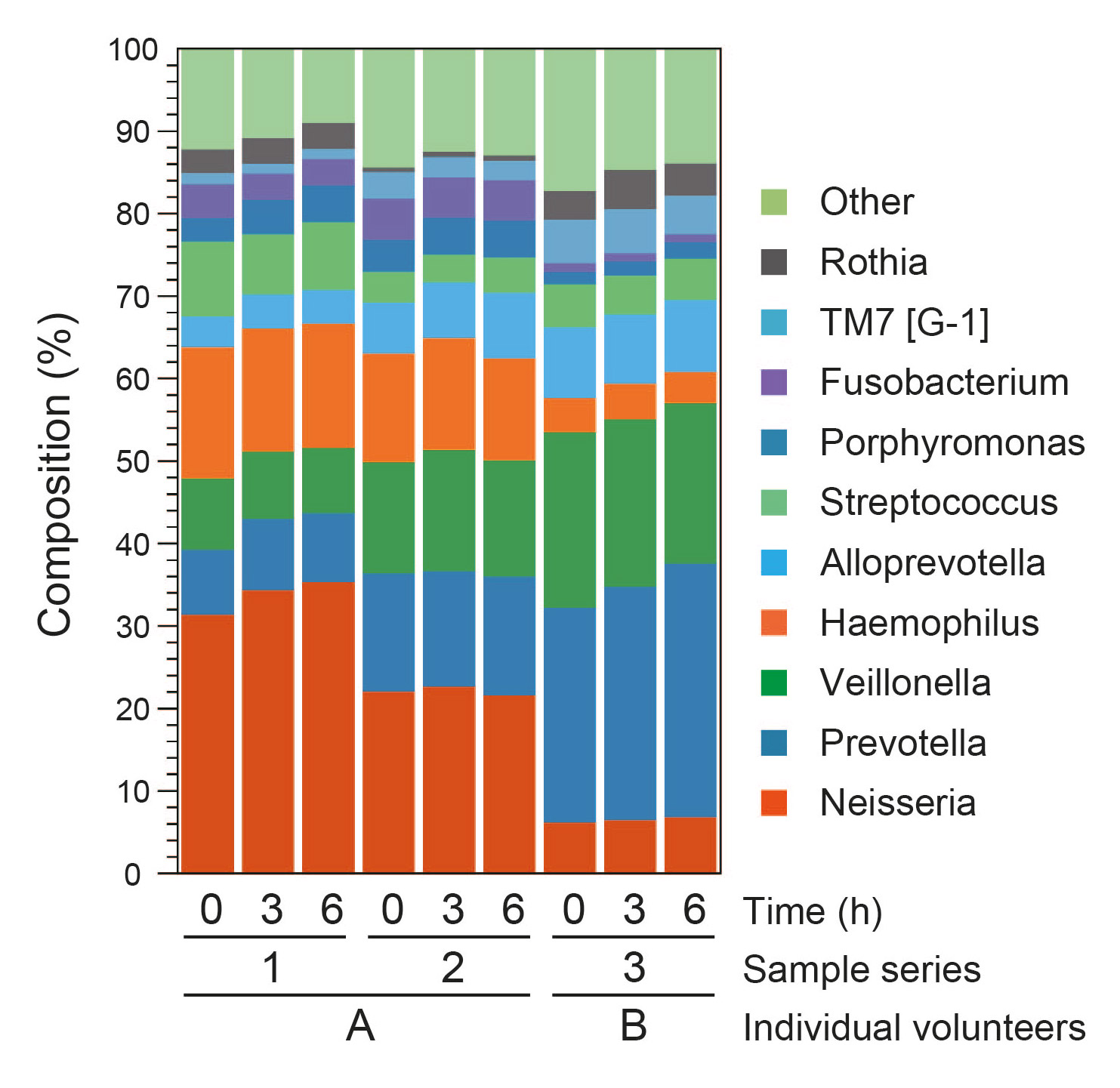
Fig. 1
Relative genus abundance of salivary samples. Three salivary specimens were collected from 2 healthy adults. The 3 samples from the left are series 1 (samples from volunteer A), the 3 samples in the middle are series 2 (samples from volunteer A), and the next 3 samples are series 3 (samples from volunteer B). The 10 most predominant genera and their relative abundance are presented.
-
Fig. 2
Relative species abundance of salivary samples. Three salivary specimens were collected from 2 healthy adults. The 3 samples from the left are series 1 (samples from volunteer A), the 3 samples in the middle are series 2 (samples from volunteer A), and the next 3 samples are series 3 (samples from volunteer B). The 10 most predominant species and their relative abundance are presented.
Results
Microbiota analysis of salivary samples
The Illumina MiSeq was performed using 9 independent samples to generate a total of 791,888 raw sequence reads (Table 1). After passing the quality filter, the number of mapped sequence reads ranged from 45,973 to 55,908 (mean ± standard error of the mean of 50,019 ± 1,160; Table 1).
Stability of the microbiota at room temperature
Figures 1 and 2 present the top 10 bacterial groups at the genus and species levels, respectively. The results of all samples revealed that the top 10 bacterial genera constitute approximately 80–90% of each sample. The most predominant members of the community belonged to the bacterial genus Neisseria in volunteer A (20–35%) and Prevotella in volunteer B (25–30%). In addition, Haemophilus, Porphyromonas and Fusobacterium were detected predominantly in volunteer A, whereas TM7 [G-1] was more predominant in volunteer B. The proportion of genus Rothia varied between sampling times in volunteer A. Although the composition of the top 10 bacterial genera was slightly fractured in all lineages for 6 h, the relative changes in each were within approximately 5%. No apparent time-dependent change in microbiota was detected in the top 10 bacterial OTUs.
Effects of room temperature storage on microbial beta diversity
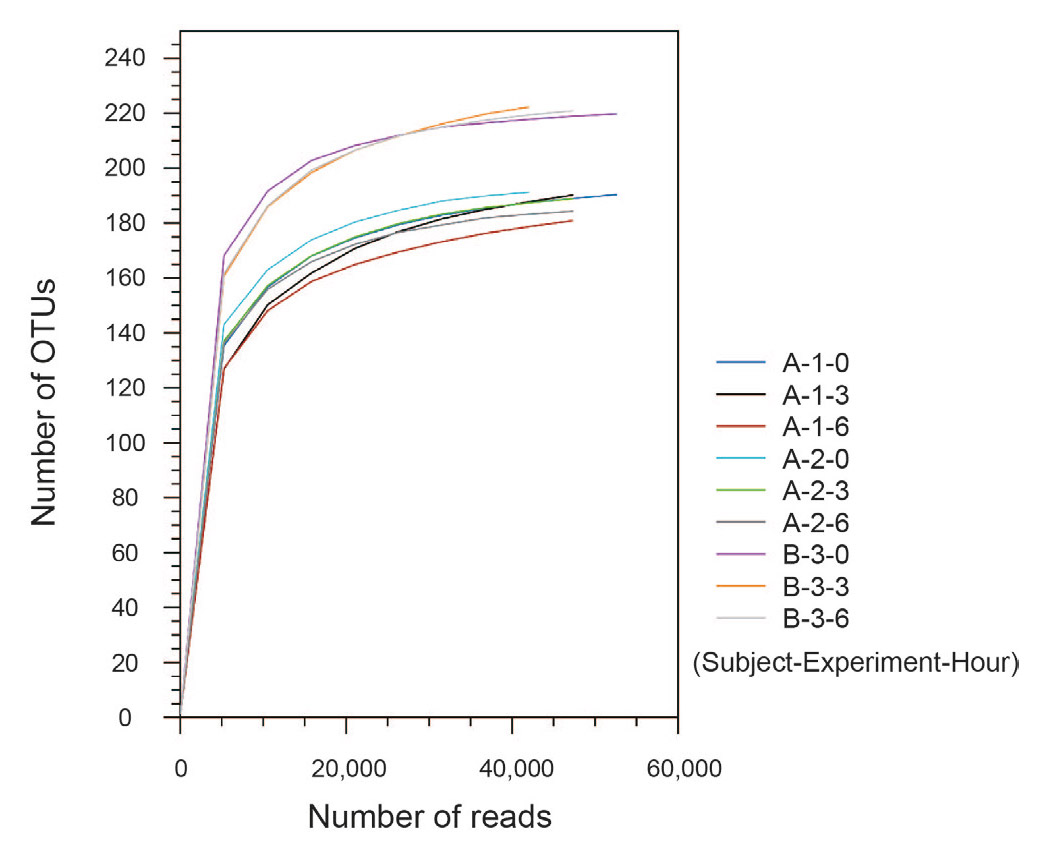
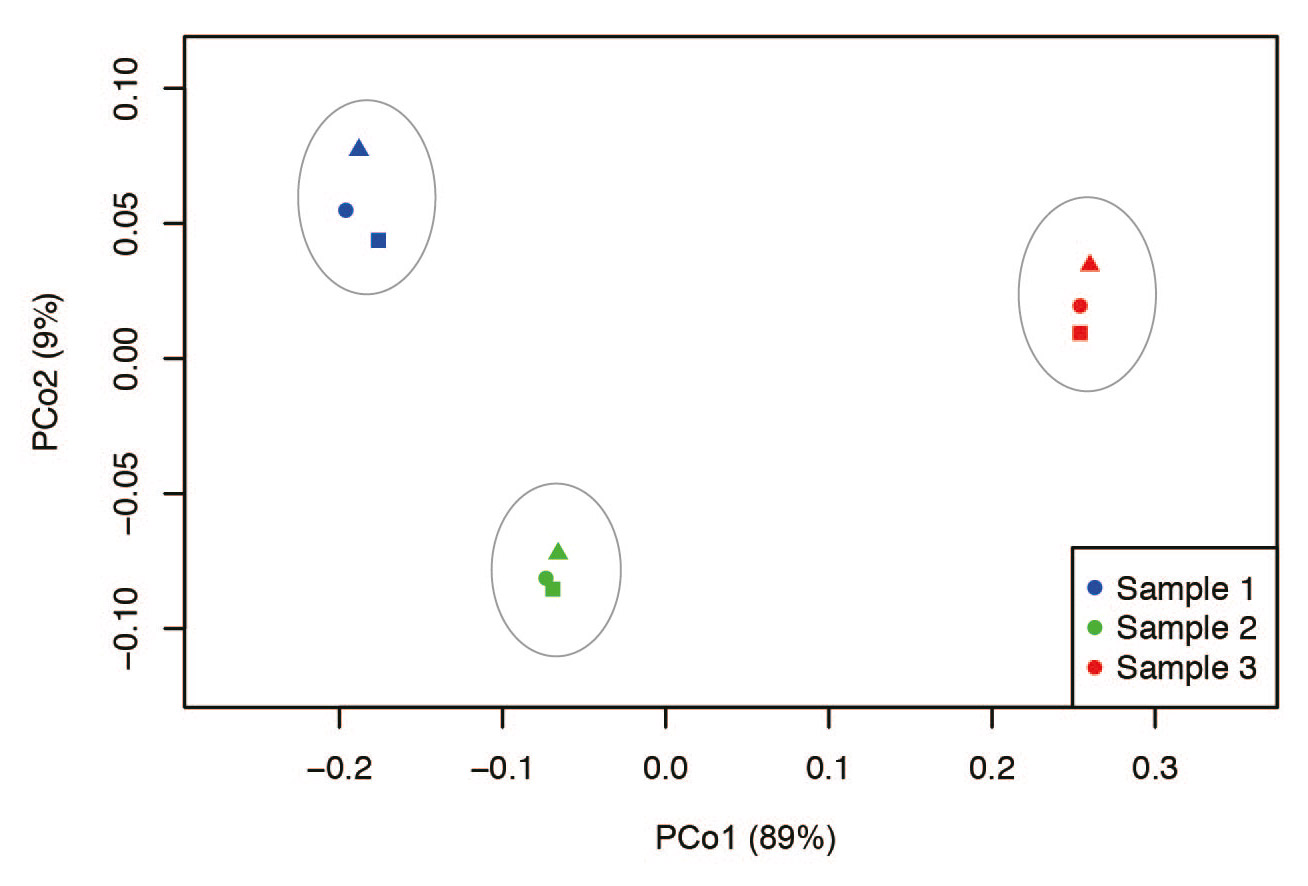
Results from rarefaction curves indicated that a sufficient number of reads was obtained for 16S rRNA analyses (Fig. 3). The PCoA plot based on the weighted UniFrac (beta diversity) metric revealed that the 9 samples clustered into 3 groups corresponding to each lineage (Fig. 4), and the difference was confirmed statistically by PERMANOVA (P < 0.01). In addition, the 2 lineage groups collected from volunteer A closely opposed each other on the PCoA plot. These results demonstrated that no overall change in community composition was observed in this experimental condition (20°C, 6 h).
-
Fig. 3
Rarefaction curves of OTUs.
-
Fig. 4
Principle coordinate analysis (PCoA) plot showing differences in bacterial community structure. Significant differences were observed at the OTU level when examined by PERMANOVA analysis of the UniFrac distance matrix (P < 0.005).
Change in salivary microbiota at the OTU levels
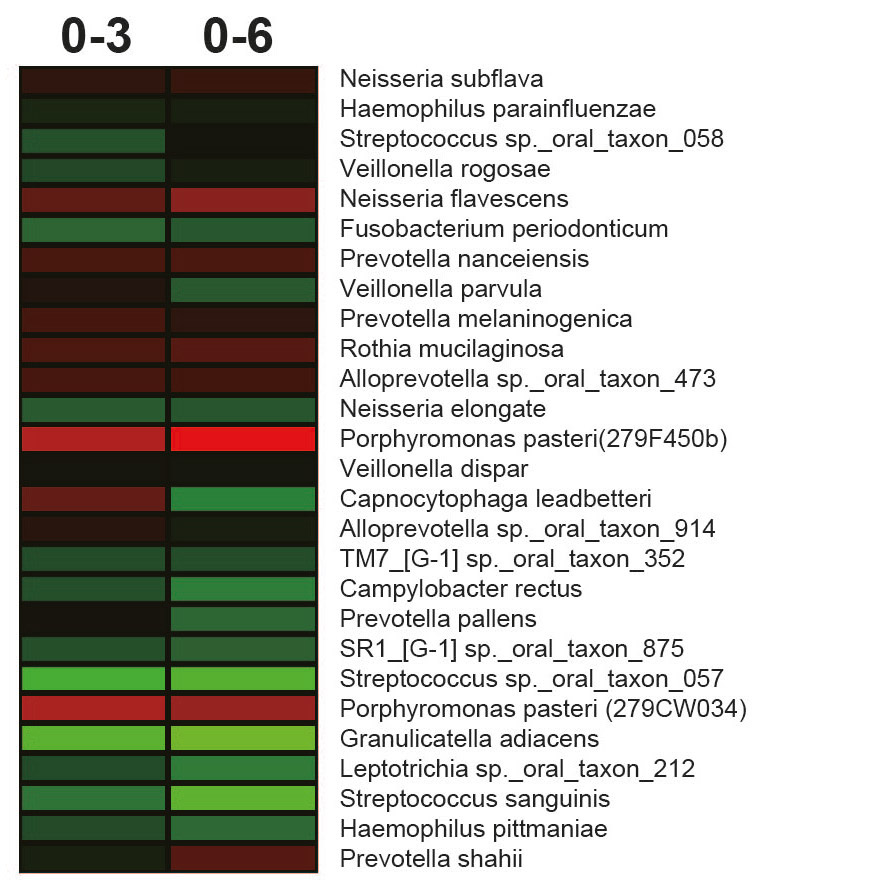
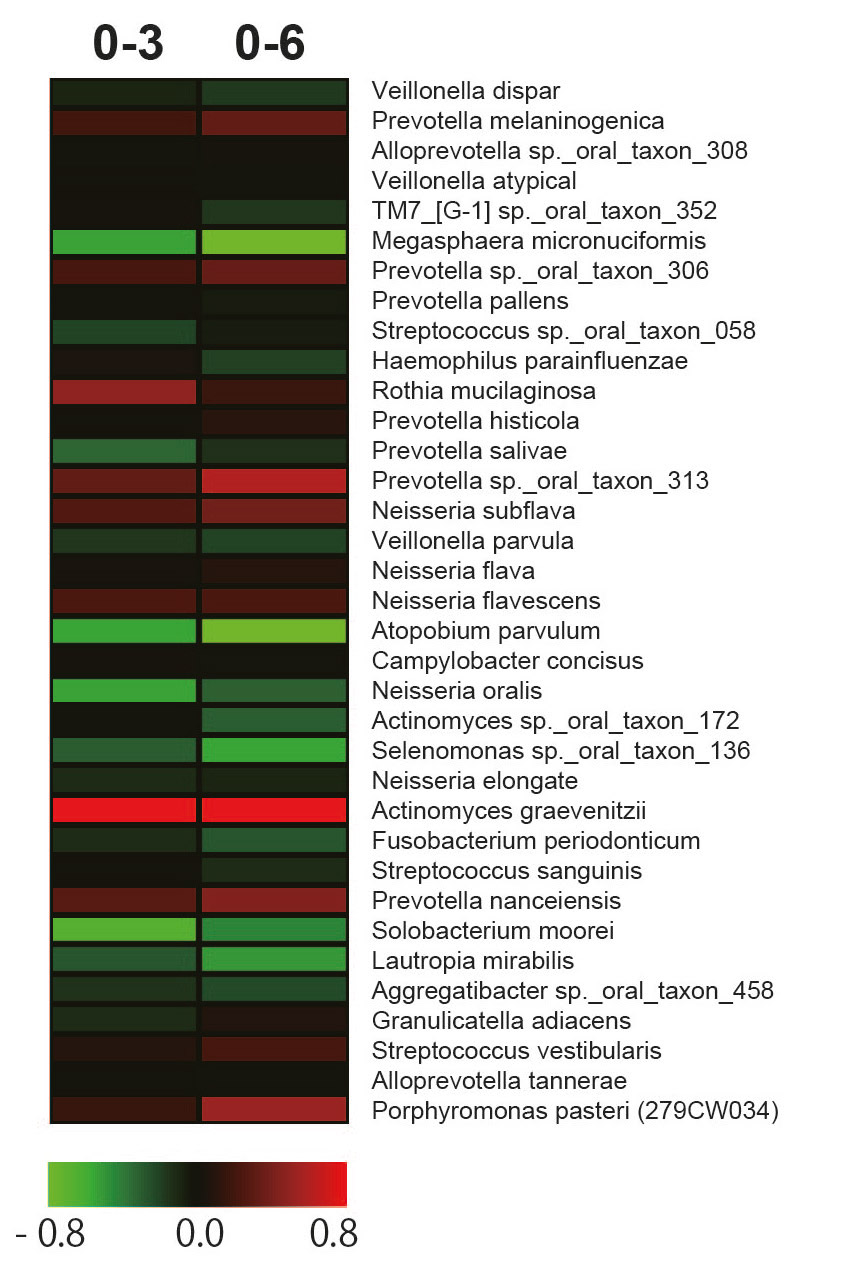
Bacterial shifts in salivary microbiota at the OTU levels associated with incubation at room temperature (20°C) were investigated and visualized as heat maps (Fig. 5). In sample 1 of volunteer A, 2 types of Porphyromonas pasteri were regarded as having increased OTUs at a 0.5 cutoff over the course of 6 h. In contrast, Granulicatella adiacens, Streptococcus sanguinis and Streptococcus sp. oral taxon 057 were decreased in the same sample. In sample 2 of volunteer A, although no OTUs were observed to have increased, Granulicatella adiacens and Selenomonas sp. were decreased. In the sample from volunteer B, Actinomyces graevenitzii, Prevotella sp. and Porphyromonas pasteri were increased, and Atopobium parvulum, Megasphaera micronuciformis and Selenomonas sp. were decreased. These results suggest that there are changes in specific bacterial taxa during room temperature storage for a few hours.
Discussion
The relationship between the indigenous bacterial flora and human health has been attracting attention, and several large-scale cohort studies are being performed, in which the duration and method of sample storage from sample collection to analysis is an issue. For fecal and vaginal microorganism-derived samples, several studies evaluating the degree of storage-induced changes in the flora have been performed, 20Kim D, Hofstaedter CE, Zhao C, Mattei L, Tanes C, Clarke E, Lauder A, Sherrill-Mix S, Chehoud C, Kelsen J, Conrad M, Collman RG, Baldassano R, Bushman FD, Bittinger K. Optimizing methods and dodging pitfalls in microbiome research. → Microbiome. 2017 May 5;5(1):52. doi: 10.1186/s40168-017-0267-5.,21Tedjo DI, Jonkers DM, Savelkoul PH, Masclee AA, van Best N, Pierik MJ, Penders J. The effect of sampling and storage on the fecal microbiota composition in healthy and diseased subjects. → PLoS One. 2015 May 29;10(5):e0126685. doi: 10.1371/journal.pone.0126685.,22Guo Y, Li SH, Kuang YS, He JR, Lu JH, Luo BJ, Jiang FJ, Liu YZ, Papasian CJ, Xia HM, Deng HW, Qiu X. Effect of short-term room temperature storage on the microbial community in infant fecal samples. → Sci Rep. 2016 May 26;6:26648. doi: 10.1038/srep26648. but no study evaluating the influence of storage conditions on oral samples has been performed. A study comparing media for oral sample storage23Luo T, Srinivasan U, Ramadugu K, Shedden KA, Neiswanger K, Trumble E, Li JJ, McNeil DW, Crout RJ, Weyant RJ, Marazita ML, Foxman B. Effects of specimen collection methodologies and storage conditions on the short-term stability of oral microbiome taxonomy. → Appl Environ Microbiol. 2016 Sep;82(18):5519–29. has been reported, but it only investigated the influence of room temperature storage for several days, and to our knowledge, no study has investigated changes over a several-hour period. In the present study, we investigated the storage conditions after salivary sample collection, aiming at securing the reliability of data collected in clinical and cohort studies on the oral flora, which are expected to be frequently performed in the future.
Regarding the study design, collected saliva was kept at around room temperature (20°C) for a specified time and changes in the bacterial flora in the sample were analyzed in DNA extracted from the sample after 3 and 6 h. More than 70,000 reads could be obtained from all 9 samples collected, and at least 45,000 reads passed through the filter. This number of reads is sufficient compared with those in other analyses.24Shi W, Qin M, Chen F, Xia B. Supragingival microbial profiles of permanent and deciduous teeth in children with mixed dentition. → PLoS One. 2016 Jan 11;11(1):e0146938. doi: 10.1371/journal.pone.0146938.,25Yu G, Phillips S, Gail MH, Goedert JJ, Humphrys MS, Ravel J, Ren Y, Caporaso NE. The effect of cigarette smoking on the oral and nasal microbiota. → Microbiome. 2017 Jan 17;5(1):3. doi: 10.1186/s40168-016-0226-6.
When time course changes in the flora were subjected to PCoA, slight changes within each sample were noted under the conditions of this study, but the characteristics of the floras observed at the time of sampling did not markedly change (Fig. 4). In the salivary samples collected with a 1-week interval from the same subject, the composition of the flora was similar, but the ratios appeared to have changed (Figs. 1 & 2). Regarding the individual OTU in recent analyses, it has been clarified that several specific OTUs markedly changed when the samples were stored at room temperature.26Amir A, McDonald D, Navas-Molina JA, Debelius J, Morton JT, Hyde E, Robbins-Pianka A, Knight R. Correcting for microbial blooms in fecal samples during room-temperature shipping. → mSystems. 2017 Mar 7;2(2). pii: e00199-16. doi: 10.1128/mSystems.00199-16. When changes were closely investigated by OTU, Porphyromonas pasteri increased in 2 of the 3 samples, whereas Selenomonas sp. oral taxon 136 and Granulicatella adiacens decreased (Fig. 5), showing that OTUs termed “bloom,” which alter during storage at room temperature, are very likely to be present in oral samples, similar to fecal samples. It is desirable to freeze samples as soon as possible when they are collected from the oral cavity of patients with periodontal disease and periimplantitis lesions in cohort and clinical sites. It is also important to know the species of OTUs that change during storage at room temperature to prepare for conditions in which such storage is difficult, in order to maintain the resolution of flora analysis data.
- Fig. 5A
- Fig. 5B
- Fig. 5C
Heat maps of the fold changes from 0 to 3 or 0 to 6 h in sample series 1 (A), 2 (B) and 3 (C). The heat maps illustrate the microbes with the highest relative abundance (> 0.5% with respect to the total number of sequences) using a binary logarithmic color scale.
This is a preliminary study and there are some limitations. First, the sample size was small. A large number of patients may be more likely to elucidate the temporal stability of salivary microbiota. Second, the observation was based on experimentation at only 1 temperature (20°C). Addressing these limitations in future studies will be important.
Conclusion
Within the limitation of the sample size in this preliminary study, it can be stated that room temperature storage of salivary samples for a few hours may not affect the overall microbiota structure, while there are some changes in specific bacterial taxa.
Acknowledgments
We thank Hisako Sakanaka for technical assistance. This work was partly supported by KAKENHI (Grants-in-Aid for Scientific Research [C] No. 16K11469, No. 16K11876 and No. 16K11877). the Japan Society for the Promotion of Science and an Osaka Dental University Oral Implant Research Grant (No. 17-02).
Competing interests
The authors declare that they have no competing interests.
Why did you conduct the research reported on in this paper?
For what reasons could others cite your paper?
How could your study’s findings have an impact on dentistry?
What is the relevance of your study’s findings to the daily practice of a dentist?
What are your recommendations for further investigation of the topic of your article?
References
| 1. | ↑ | Sender R, Fuchs S, Milo R. Are we really vastly outnumbered? Revisiting the ratio of bacterial to host cells in humans. → Cell. 2016 Jan;164(3):337–40. |
| 2. | ↑ | Thomas S, Izard J, Walsh E, Batich K, Chongsathidkiet P, Clarke G, Sela DA, Muller AJ, Mullin JM, Albert K, Gilligan JP, DiGuilio K, Dilbarova R, Alexander W, Prendergast GC. The host microbiome regulates and maintains human health: a primer and perspective for non-microbiologists. → Cancer Res. 2017 Apr;77(8):1783–812. |
| 3. | ↑ | Schroeder BO, Bäckhed F. Signals from the gut microbiota to distant organs in physiology and disease. → Nat Med. 2016 Oct;22(10):1079–89. |
| 4. | ↑ | Hong SW, Kim KS, Surh CD. Beyond hygiene: commensal microbiota and allergic diseases. → Immune Netw. 2017 Feb;17(1):48–59. |
| 5, 6. | ↑ | Krishnan K, Chen T, Paster BJ. A practical guide to the oral microbiome and its relation to health and disease. → Oral Dis. 2017 Apr;23(3):276–86. |
| 7. | ↑ | Koch CD, Gladwin MT, Freeman BA, Lundberg JO, Weitzberg E, Morris A. Enterosalivary nitrate metabolism and the microbiome: intersection of microbial metabolism, nitric oxide and diet in cardiac and pulmonary vascular health. → Free Radic Biol Med. 2017 Apr;105:48–67. |
| 8. | ↑ | Aas JA, Paster BJ, Stokes LN, Olsen I, Dewhirst FE. Defining the normal bacterial flora of the oral cavity. → J Clin Microbiol. 2005 Nov;43(11):5721–32. |
| 9. | ↑ | Zaura E. Next-generation sequencing approaches to understanding the oral microbiome. → Adv Dent Res. 2012 Sep;24(2):81–5. |
| 10. | ↑ | Yamagishi J, Sato Y, Shinozaki N, Ye B, Tsuboi A, Nagasaki M, Yamashita R. Comparison of boiling and robotics automation method in DNA extraction for metagenomic sequencing of human oral microbes. → PLoS One. 2016 Apr;11(4):e0154389. |
| 11, 14. | ↑ | Abusleme L, Hong BY, Dupuy AK, Strausbaugh LD, Diaz PI. Influence of DNA extraction on oral microbial profiles obtained via 16S rRNA gene sequencing. → J Oral Microbiol. 2014 Apr 23;6. doi: 10.3402/jom. v6.23990. |
| 12. | ↑ | Lazarevic V, Gaïa N, Girard M, François P, Schrenzel J. Comparison of DNA extraction methods in analysis of salivary bacterial communities. → PLoS One. 2013 Jul 3;8(7):e67699. doi: 10.1371/journal.pone.0067699. |
| 13. | ↑ | Vesty A, Biswas K, Taylor MW, Gear K, Douglas RG. Evaluating the impact of DNA extraction method on the representation of human oral bacterial and fungal communities. → PLoS One. 2017 Jan 18;12(1):e0169877. doi: 10.1371/journal.pone.0169877. |
| 15, 19, 23. | ↑ | Luo T, Srinivasan U, Ramadugu K, Shedden KA, Neiswanger K, Trumble E, Li JJ, McNeil DW, Crout RJ, Weyant RJ, Marazita ML, Foxman B. Effects of specimen collection methodologies and storage conditions on the short-term stability of oral microbiome taxonomy. → Appl Environ Microbiol. 2016 Sep;82(18):5519–29. |
| 16, 18, 20. | ↑ | Kim D, Hofstaedter CE, Zhao C, Mattei L, Tanes C, Clarke E, Lauder A, Sherrill-Mix S, Chehoud C, Kelsen J, Conrad M, Collman RG, Baldassano R, Bushman FD, Bittinger K. Optimizing methods and dodging pitfalls in microbiome research. → Microbiome. 2017 May 5;5(1):52. doi: 10.1186/s40168-017-0267-5. |
| 17, 21. | ↑ | Tedjo DI, Jonkers DM, Savelkoul PH, Masclee AA, van Best N, Pierik MJ, Penders J. The effect of sampling and storage on the fecal microbiota composition in healthy and diseased subjects. → PLoS One. 2015 May 29;10(5):e0126685. doi: 10.1371/journal.pone.0126685. |
| 22. | ↑ | Guo Y, Li SH, Kuang YS, He JR, Lu JH, Luo BJ, Jiang FJ, Liu YZ, Papasian CJ, Xia HM, Deng HW, Qiu X. Effect of short-term room temperature storage on the microbial community in infant fecal samples. → Sci Rep. 2016 May 26;6:26648. doi: 10.1038/srep26648. |
| 24. | ↑ | Shi W, Qin M, Chen F, Xia B. Supragingival microbial profiles of permanent and deciduous teeth in children with mixed dentition. → PLoS One. 2016 Jan 11;11(1):e0146938. doi: 10.1371/journal.pone.0146938. |
| 25. | ↑ | Yu G, Phillips S, Gail MH, Goedert JJ, Humphrys MS, Ravel J, Ren Y, Caporaso NE. The effect of cigarette smoking on the oral and nasal microbiota. → Microbiome. 2017 Jan 17;5(1):3. doi: 10.1186/s40168-016-0226-6. |
| 26. | ↑ | Amir A, McDonald D, Navas-Molina JA, Debelius J, Morton JT, Hyde E, Robbins-Pianka A, Knight R. Correcting for microbial blooms in fecal samples during room-temperature shipping. → mSystems. 2017 Mar 7;2(2). pii: e00199-16. doi: 10.1128/mSystems.00199-16. |








Leave a Reply
Be the First to Comment!